CONDENA MH CLÚSTER ATAQUE ARMADO A HOSPITAL ARCÁNGELES
Leer más
Off balance: Interferons in COVID-19 lung infections
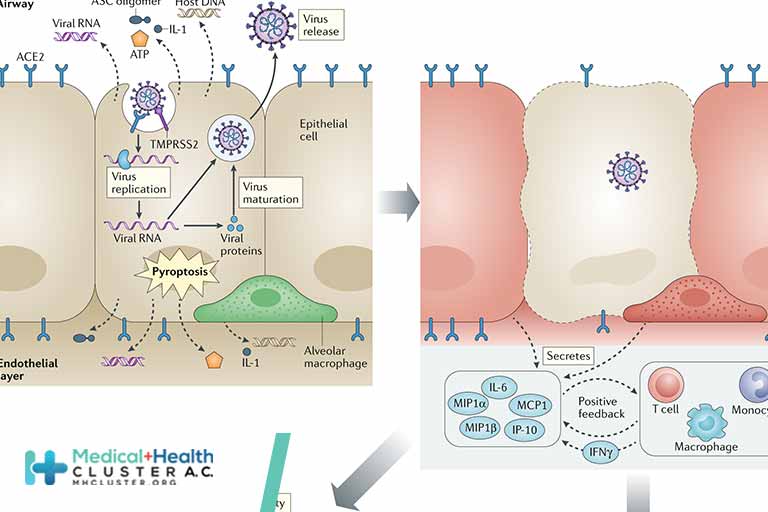
Interferons are innate and adaptive cytokines involved in many biological responses, in particular, viral infections. With the final response the result of the balance of the different types of Interferons. Cytokine storms are physiological reactions observed in humans and animals in which the innate immune system causes an uncontrolled and excessive release of pro-inflammatory signaling molecules. The excessive and prolonged presence of these cytokines can cause tissue damage, multisystem organ failure and death. The role of Interferons in virus clearance, tissue damage and cytokine storms are discussed, in view of COVID-19 caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The imbalance of Type I, Type II and Type III Interferons during a viral infection contribute to the clinical outcome, possibly together with other cytokines, in particular, TNFα, with clear implications for clinical interventions to restore their correct balance.
Interferons (IFNs) are essential cytokines that mediate the host resistance response to viral infections. In the absence of IFNs or compromised cellular programs induced by IFNs, vertebrates may be susceptible to lethal viral infections. There are 21 human IFNs, classified in 3 types (16 Type-I: 12 IFNαs, IFNβ, IFNε, IFNκ, and IFNω; 1 Type-II, IFNγ; and 4 Type-III, IFNλ1, IFNλ2, IFNλ3, and IFNλ4), involved in innate and adaptive immune responses against not only to viruses but also to other pathogens as well as to tumors and in tissue repair. In general, the production of distinct IFNs depends on the cell type and is stimulated during viral infection when the viral nucleic acid in the cytoplasm is detected by intracellular pattern recognition receptors (PRRs). Then, the produced and secreted IFNs will bind and activates heterodimeric receptors (IFNRs) on the cells in autocrine and paracrine mechanisms that initiate a complex cascade of signaling, culminating in transcription of hundreds of different Interferon-Stimulated Genes (ISGs) resulting in strong antiviral innate and adaptive immune responses Briefly, Type I and III IFNs bind to their heterodimeric receptors activating the JAK/STAT pathway, which in turn activate the heterotrimeric transcription factor complex ISGF3, which consists of phosphorylated STAT1/STAT2 and IRF9. However, unphosphorylated STAT molecules and IRF9-STAT2 homodimers have also been described [
[4]
]. Type II IFN also signals through the JAK/STAT pathway, resulting in the formation of phosphorylated STAT1 homodimers, also known as IFNγ-activated factor (GAF). Activated ISGF3 and GAF, resulting respectively from type I/III and type II IFN actions, translocate to the nucleus and bind IFN-stimulated response elements (ISRE) and gamma-activated sequences (GAS), respectively, in the promoter regions of ISGs (Figure 1). In this review, we will discuss the recent data showing the association of COVID19 severity with the correct balance between the IFN types and other cytokines for a favorable clinical outcome during infection by SARS-CoV-2.

Figure 1– Type I, II and III Interferons, their receptors and signalings. (A) The Type I, II and III IFN ligands and their receptors; (B) Kinases associated with the Type I, II and III receptors that mediate IFN signallings through receptor activation by ligand binding. Jak1 (Janus Kinase 1) interacts with STAT1 (Signal Transducer and Activator of Transcription 1) as well as to IFNRA1 (Interferon receptor alpha 1). Tyk2 (Tyrosine kinase 2) interacts to STAT2 as well as to IFNRA2. IFNRA1 forms a heterodimer with IFNRA2 that binds with Type I IFN; Jak1 and STAT1 also interacts to IFNLR1. IL-10RB (Interleukin-10 receptor B) also interacts with Tyk2 and STAT2. IFNLR1 forms a heterodimer with IL-10RB that binds to Type III IFNs. Jak1 and STAT1 also interacts to IFNGR1 and JAK2 and STAT1 also interacts with IFNGR2. IFNGR1 forms a heterodimer with IFNGR2 that binds to Type II IFNs; (C) Upon ligand binding and activation, the STAT1 or STAT2 are phosphorylated and the STAT1 homodimers go to the nucleous and activate the transcription of genes with the GAS (gama interferon activation sites) promoters; STAT1 and STAT2 heterodimers binds to IRF9 (interferon regulatory factor 9) and the complex goes to the nucleous and activates the the transcription of genes with the ISRE (Interferon-stimulated responsive element) promoters. Some commom ISGs (trafd1, TNF receptor-associated factor -type zinc finger domain containing 1; parp 9, Poly(ADP-ribose) polymerase family member 9; parp12, Poly(ADP-ribose) polymerase family member 12; parp14, Poly(ADP-ribose) polymerase family member 14; nmi, N-myc and STAT interactor; epsti1, epithelial stromal interaction 1; eif2ak2, eukaryotic translation initiation factor 2 alpha kinase 2; irf1, interferon regulatory factor 1; irf7, interferon regulatory factor 7; irf8, interferon regulatory factor 8; tmem67, transmembrane protein 67; tmem140, transmembrane protein 140; tmem173, transmembrane protein 173; dtx3l, deltex E3 ubiquitin-protein ligase; samhd1, SAM (sterile alpha-motif) and HD (histidine-aspartate) domain containing deoxynucleoside triphosphate truphosphohydrolase 1; stat1, signal transducer and activator of transcription 1; cd274, cluster of differentiation 274) induced by Type I, II and III IFNs are indicated by * [
Créditos: Comité científico Covid




